Biological Langauge Models
Autoregressive generative model for functional synthetic yeast promoter sequence design
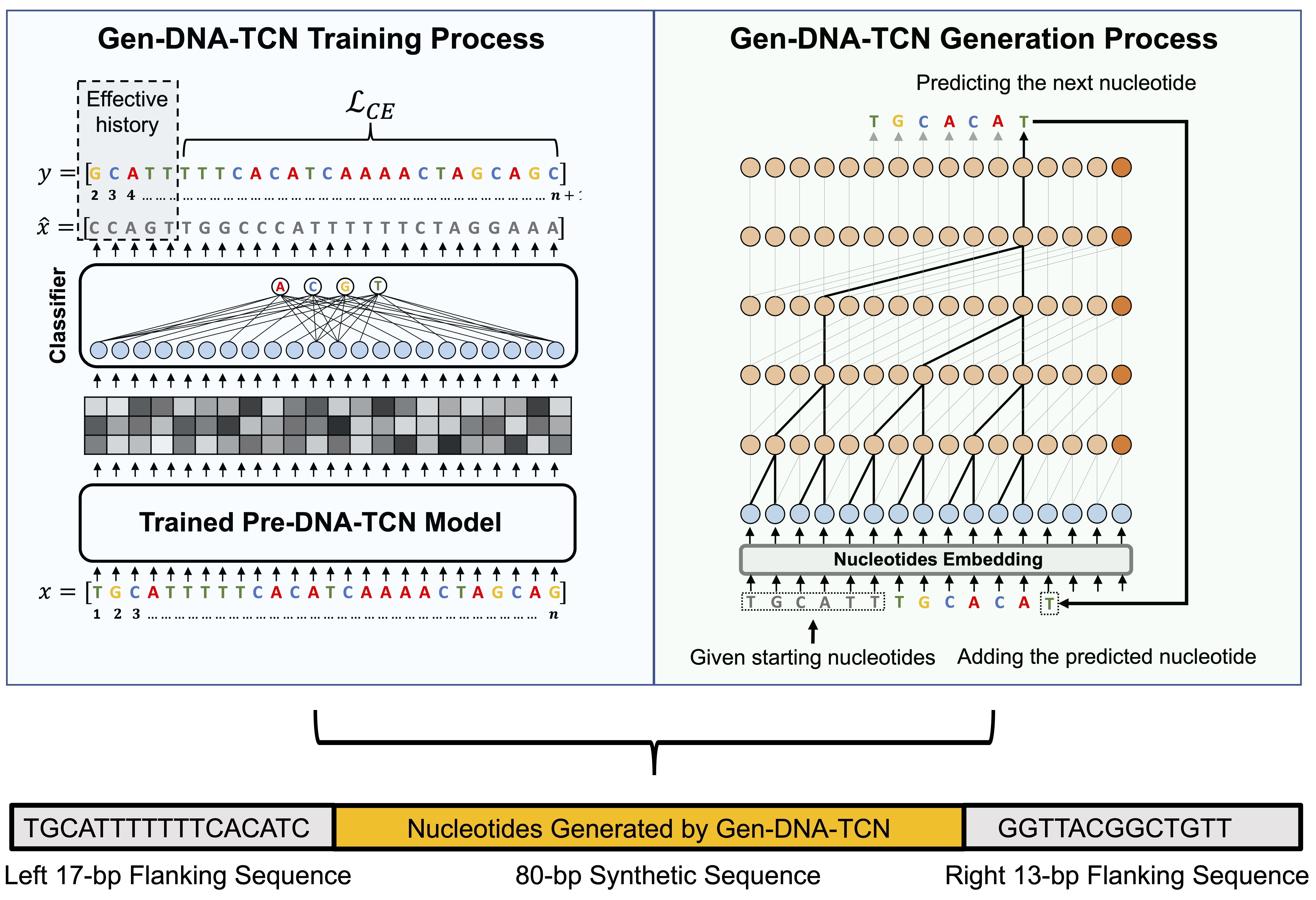
Source code: https://github.com/ibrahimsaggaf/Gen-DNA-TCN
Pre-trained models: https://zenodo.org/records/18182470
Citation: Alsaggaf, I., Freitas, A.A., Magalhaes, J.P. and Wan, C. (2026) Functional yeast promoter sequence design using autoregressive generative models, bioRxiv. DOI: 10.1101/2024.10.22.619701.
Autoregressive predictive model for estimating yeast gene expression profiles
Source code: https://github.com/de-Boer-Lab/random-promoter-dream-challenge-2022/tree/main/dream_submissions/Wan%26Barton_BBK/code
Citation: Rafi, A.M., et al. (2024) A community effort to optimize sequence-based deep learning models of gene regulation, Nature Biotechnology.
Alsaggaf, I., et al. (2022) Team WanBarton_BBK Submission. Available at: https://github.com/de-Boer-Lab/random-promoter-dream-challenge-2022/blob/main/dream_submissions/Wan%26Barton_BBK/report.pdf
Contrastive Learning
Augmentation-free single-cell RNA-Seq contrastive learning (AF-RCL)

Source code: https://doi.org/10.6084/m9.figshare.28830311.v1
Pre-trained models: https://doi.org/10.5281/zenodo.15109736
Citation: Alsaggaf, I., Buchan, D. and Wan, C. (2025) Less is more: Improving cell-type identification with augmentation-free single-cell RNA-Seq contrastive learning, Bioinformatics.
Enhanced Gaussian noise augmentation-based protein-protein interaction network embedding contrastive learning (EGsCL)
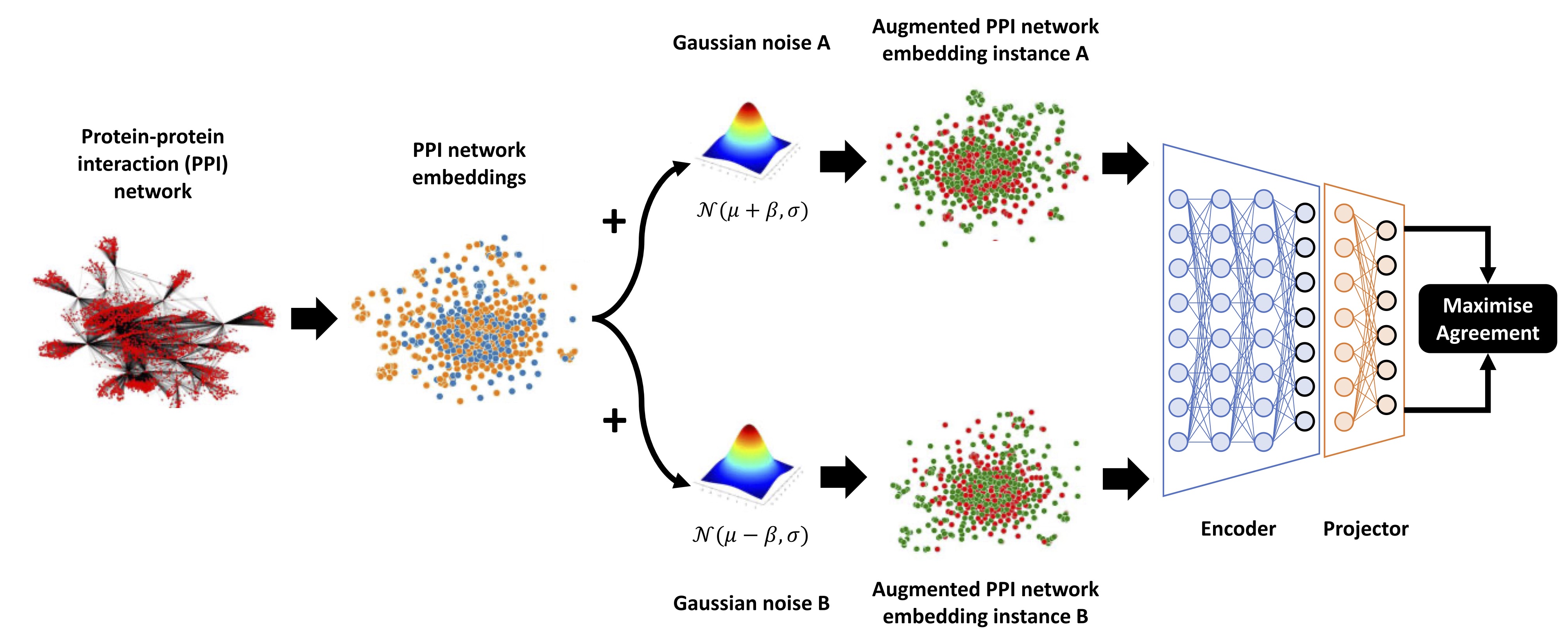
Source code: https://github.com/ibrahimsaggaf/EGsCL
Pre-trained models: https://doi.org/10.5281/zenodo.12143797
Citation: Alsaggaf, I., Freitas, A.A. and Wan, C. (2024) Predicting the pro-longevity or anti-longevity effect of model organism genes with enhanced Gaussian noise augmentation-based contrastive learning on protein–protein interaction networks, NAR Genomics and Bioinformatics.
Gaussian noise augmentation-based single-cell RNA-Seq contrastive learning (GsRCL)
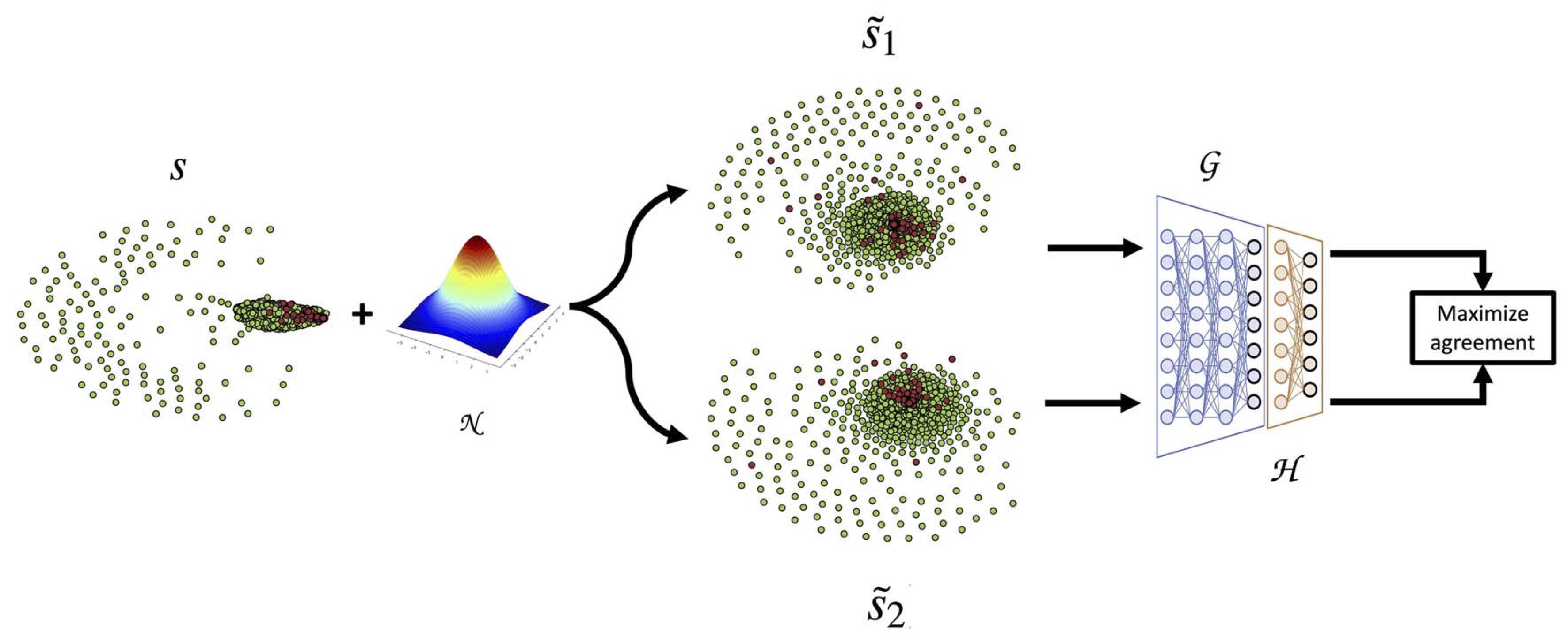
Academic server: https://bioinf.cs.ucl.ac.uk/psipred
Source code: https://github.com/ibrahimsaggaf/GsRCL
Pre-trained models: https://doi.org/10.5281/zenodo.10050548.
Citation: Alsaggaf, I., Buchan, D. and Wan, C. (2024) Improving cell-type identification with Gaussian noise-augmented single-cell RNA-seq contrastive learning, Briefings in Functional Genomics, elad059.